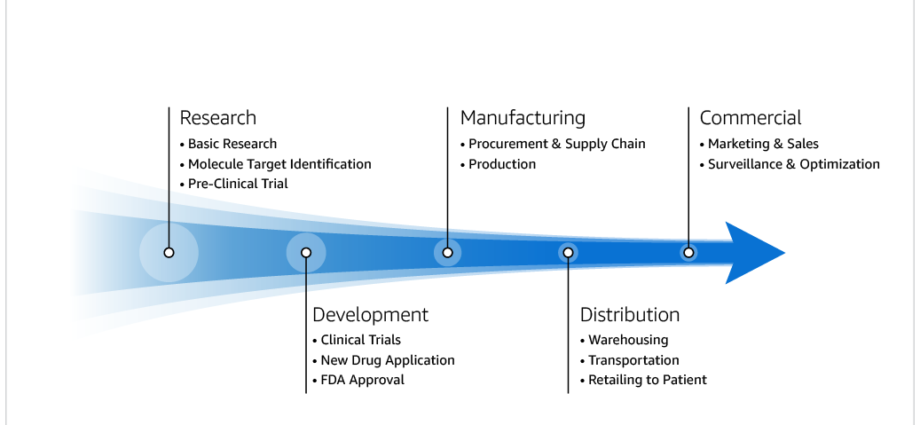
It takes biopharma companies over 10 years, at a cost of over $2 billion and with a failure rate of over 90%, to deliver a new drug to patients. The Market to Molecule (M2M) value stream process, which biopharma companies must apply to bring new drugs to patients, is resource-intensive, lengthy, and highly risky. Nine out of ten biopharma companies are AWS customers, and helping them streamline and transform the M2M processes can help deliver drugs to patients faster, reduce risk, and bring value to our customers.
Pharmaceutical companies are taking a new approach to drug discovery, looking for variants in the human genome and linking them to diseases. This genetic validation approach can improve the success ratio in the M2M value stream process by focusing on the root cause of disease and the gene variants.
As depicted in the following M2M value stream diagram, the Research process (and the Basic Research sub-process) is critical to downstream processes where linking the gene variant to a disease occurs, and is instrumental in defining the target molecule. This can be a critical step in expediting and reducing the cost of delivering a new drug to patients.

To transform the M2M value stream process, our customer has been working on associating genes with diseases by using their large dataset of over 2 million sequenced exomes (genes that are expressed into proteins). To accomplish this, the customer’s clinical scientists have to develop methods to navigate through the enormous dataset by using online genome browsers, a mechanical data-first experience that doesn’t fully meet the needs of users. Starting with a search query to get results, the typical interactions of navigating levels, filtering, waiting, and repeating the search can be time-consuming and tedious. Simplifying the UI from the traditional human browser to a conversational AI assistant can enhance the user experience in the clinical research process.
Generative AI is a promising next step in the evolutionary process of leading this change. As generative AI started to make significant impact in healthcare and life sciences, this use case was primed for generative AI experimentation. In collaboration with the customer, AWS built a custom approach of posting a question or a series of questions, allowing scientists to have more flexibility and agility for exploring the genome. Our customer aimed at saving researchers countless hours of work using a new generative AI-enabled gene assistant. By asking a question, or a series of questions, scientists have more flexibility and agility in exploring the genome. Identifying variants and their potential correlation with diseases can be done more efficiently using words, rather than filters, settings, and buttons. With a more streamlined research process, we can help increase the likelihood of leading to new breakthroughs.
This post explores deploying a text-to-SQL pipeline using generative AI models and Amazon Bedrock to ask natural language questions to a genomics database. We demonstrate how to implement an AI assistant web interface with AWS Amplify and explain the prompt engineering strategies adopted to generate the SQL queries. Finally, we present instructions to deploy the service in your own AWS account. Amazon Bedrock is a fully managed service that provides access to large language models (LLMs) and other foundation models (FMs) from leading AI companies through a single API, allowing you to use it instantly without much effort, saving developers valuable time. We used the AWS HealthOmics variant stores to store the Variant Call Format (VCF) files with omics data. A VCF file is typically the output of a bioinformatics pipeline. VCFs encode Single Nucleotide Polymorphisms (SNPs) and other structural genetic variants. The format is further described on the 1000 Genomes project website. We used the AWS HealthOmics – End to End workshop to deploy the variants and annotation stores.
Although this post focuses on a text-to-SQL approach to an omics database, the generative AI approaches discussed here can be applied to a variety of complex schemas of relational databases.
Text-to-SQL for genomics data
Text-to-SQL is a task in natural language processing (NLP) to automatically convert natural language text into SQL queries. This involves translating the written text into a structured format and using it to generate an accurate SQL query that can run on a database. The task is difficult because there are big differences between human language, which is flexible, ambiguous, and dependent on context, and SQL, which is structured.
Before LLMs for text-to-SQL, user queries had to be preprocessed to match specific templates, which were then used to rephrase the queries. This approach was use case-specific and required data preparation and manual work. Now, with LLMs, the text-to-SQL task has undergone a major transformation. LLMs continue to showcase key performance improvements in generating valid SQL queries from natural language queries. Relying on pre-trained models trained on massive datasets, LLMs can identify the relationships between words in language and accurately predict the next ones to be used.
However, although LLMs have remarkable performance in many text-to-SQL problems, they have limitations that lead to hallucinations. This post describes the main approaches used to overcome these limitations.
There are two key strategies to achieve high accuracy in text-to-SQL services:
- Prompt engineering – The prompt is structured to annotate different components, such as pointing to columns and schemas, and then instructing the model on which type of SQL to create. These annotations act as instructions that guide the model in formatting the SQL output correctly. For example, a prompt might contain annotations showing specific table columns and guiding the model to generate a SQL query. This approach allows for more control over the model’s output by explicitly specifying the desired structure and format of the SQL query.
- Fine-tuning – You can start with a pre-trained model on a large general text corpus and then proceed with an instruction-based fine-tuning with labeled examples to improve the model’s performance on text-to-SQL tasks. This process adapts the model to the target task by directly training it on the end task, but it requires a substantial number of text-SQL examples.
This post focuses on the prompt engineering strategy for SQL generation. AWS customers deploy prompt engineering strategies first because they’re efficient in returning high-quality results and require a less complex infrastructure and process. For more details and best practices on when to follow each approach, refer to Best practices to build generative AI applications on AWS.
We experimented with prompt engineering using chain-of-thought and tree-of-thought approaches to improve the reasoning and SQL generation capabilities. The chain-of-thought prompting technique guides the LLMs to break down a problem into a series of intermediate steps or reasoning steps, explicitly expressing their thought process before arriving at a definitive answer or output.
Using prompts, we compelled the LLM to generate a series of statements about its own reasoning, allowing the LLM to articulate its reasoning process to produce accurate and understandable outputs. The tree-of-thought approach introduces a structured branching approach to the reasoning process. Instead of a linear chain, we prompt the LLM to generate a tree-like structure, where each node represents a sub-task, sub-question, or intermediate step in the overall problem-solving process.
Solution Overview
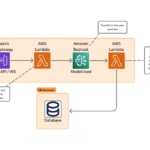
The following architecture depicts the solution and AWS services we used to accomplish the prototype.

The workflow consists of the following steps:
- A scientist submits a natural language question or request to a chat web application connected through Amplify and integrated with an AWS AppSync GraphQL API.
- The request is submitted to Amazon API Gateway, which transfers the request to an AWS Lambda function that contains the text-to-SQL implementation. We recommend the implementation of a second helper Lambda function to fetch variants data, or gene names, or ClinVar listed diseases, to simplify the user experience and facilitate the SQL generation process.
- The text-to-SQL Lambda function receives the natural language request, merges the input question with the prompt template, and submits to Amazon Bedrock to generate the SQL.
- Our implementation also adds a step to simplify the incoming history into a single request. We submit a request to Amazon Bedrock to transform the historical inputs from that user session into a simplified natural language request. This step is optional.
- With the generated SQL, the Lambda function submits the query to Amazon Athena to retrieve the genomic data from the Amazon Simple Storage Service (Amazon S3) bucket.
- If successful, the Lambda function updates the user session stored in Amazon DynamoDB through an AWS AppSync request. That change will automatically appear on the UI that is subscribed to changes to the session table.
- If an error occurs, the code attempts to re-generate the SQL query, passing the returned error as input and requesting it to fix the error. The Lambda function then reruns the re-generated SQL against Athena and returns the result.
Generative AI approaches to text-to-SQL
We tested the following prompt-engineering strategies:
- LLM SQL agents
- LLM with Retrieval Augmented Generation (RAG) to detect tables and columns of interest
- Prompt engineering with full description of tables and columns of interest
- Prompt engineering with chain-of-thought and tree-of-thought approaches
- Prompt engineering with a dynamic few-shot approach
We didn’t achieve good results with SQL agents. We experimented with LangChain SQL agents. It was difficult for the agent to use contextual information from the dataset to generate accurate and syntactically correct SQL. A big challenge in omics data is that certain columns are arrays of structs or maps. At the time of building this project, the agents were incapable of detecting these nuances and failed to generate relevant SQL.
We experimented with a RAG approach to retrieve relevant tables and columns, given a user question. Then we informed the LLM by prompting it to generate a SQL query using only those tables and columns. A motivation behind this experiment is that a RAG approach can deal well with hundreds or thousands of columns or tables. However, this approach also didn’t return good results. This RAG approach returned too many irrelevant variables to be used in each SQL generation.
The next three approaches were successful, and we used them in combination to get the highest accuracy on synthetically correct SQL generation.
A first prompt idea we tested was to provide a full description of the main tables and columns to be used in the SQL generation given a user question. In the following example, we show a snapshot of the prompts used to describe the 1000 Genome variants tables. The goal of the prompt with database tables and column descriptions is to teach the LLM how to use the schema to generate queries. We approached it as if teaching a new developer that will write queries to that database, with examples of SQL queries to extract the correct dataset, how to filter the data, and only using the most relevant columns.
The team also worked with the creation of a prompt that used the concept of chain-of-thought and its evolution tree-of-thought to improve the reasoning and SQL generation capabilities.
The chain-of-thought prompting technique encourages LLMs to break down a problem into a series of intermediate steps, explicitly expressing their thought process before arriving at a definitive answer or output. This approach takes inspiration from the way humans often break down problems into smaller, manageable parts.
Through the use of prompts, we compelled the LLM to generate a chain-of-thought, letting the LLM articulate its reasoning process and produce more accurate and understandable outputs. This technique has the potential to improve performance on tasks that require multi-step reasoning, such as SQL generation from open-ended natural language questions. This approach presented excellent results with the FM that we tested.
As a next step in our experimentation, we used the tree-of-thought technique to generate even better results than the chain-of-thought approach. The tree-of-thought approach introduces a more structured and branching approach to the reasoning process. Instead of a linear chain, we prompt the LLM to generate a tree-like structure, where each node represents a sub-task, sub-question, or intermediate step in the overall problem-solving process. The following example presents how we used these two approaches in the prompt template:
Finally, we tested a few-shot and a dynamic few-shot approach. The few-shot approach is a prompting technique used in prompt engineering for LLMs. It involves providing the LLM with a few examples or demonstrations, along with the input prompt, to guide the model’s generation or output. In the few-shot setting, the prompt comprises the following:
- An instruction or task description
- A few examples or demonstrations of the desired output, given a specific input
- The new input for which the LLM will generate an output
By exposing the LLM to these examples, the model recognizes better patterns and infers the underlying rules or mappings between the input and desired output.
The dynamic few-shot approach extends the few-shot prompting technique. It introduces the concept of dynamically generating or selecting the examples or demonstrations used in the prompt, based on the specific input or context. In this approach, instead of providing a fixed set of examples, the prompt generation process involves:
- Analyzing the input or context
- Creating embeddings of the examples and of the input, and retrieving or generating relevant examples or demonstrations tailored to the specific input by applying a semantic search
- Constructing the prompt with the selected examples and the input
Conclusion
This post demonstrated how to implement a text-to-SQL solution to democratize the access to omics data for users that aren’t data analytics specialists. The approach used HealthOmics and Amazon Bedrock to generate SQL based on natural language queries. This approach has the potential to provide access to omics data to a larger audience than what is available today.
The code is available in the accompanying GitHub repo. The deployment instructions for the HealthOmics variants and annotation store can be found in the AWS HealthOmics – End to End workshop. The deployment instructions for the text-to-SQL project are available in the README file.
We would like to acknowledge Thomaz Silva and Saeed Elnaj for their contributions to this blog. It couldn’t have been done without them.
About the Authors
 Ganesh Raam Ramadurai is a Senior Technical Program Manager at Amazon Web Services (AWS), where he leads the PACE (Prototyping and Cloud Engineering) team. He specializes in delivering innovative, AI/ML and Generative AI-driven prototypes that help AWS customers explore emerging technologies and unlock real-world business value. With a strong focus on experimentation, scalability, and impact, Ganesh works at the intersection of strategy and engineering—accelerating customer innovation and enabling transformative outcomes across industries.
Ganesh Raam Ramadurai is a Senior Technical Program Manager at Amazon Web Services (AWS), where he leads the PACE (Prototyping and Cloud Engineering) team. He specializes in delivering innovative, AI/ML and Generative AI-driven prototypes that help AWS customers explore emerging technologies and unlock real-world business value. With a strong focus on experimentation, scalability, and impact, Ganesh works at the intersection of strategy and engineering—accelerating customer innovation and enabling transformative outcomes across industries.
 Jeff Harman is a Senior Prototyping Architect on the Amazon Web Services (AWS) Prototyping and Cloud Engineering team, he specializes in developing innovative solutions that leverage AWS’s cloud infrastructure to meet complex business needs. Jeff Harman is a seasoned technology professional with over three decades of experience in software engineering, enterprise architecture, and cloud computing. Prior to his tenure at AWS, Jeff held various leadership roles at Webster Bank, including Vice President of Platform Architecture for Core Banking, Vice President of Enterprise Architecture, and Vice President of Application Architecture. During his time at Webster Bank, he was instrumental in driving digital transformation initiatives and enhancing the bank’s technological capabilities. He holds a Master of Science degree from the Rochester Institute of Technology, where he conducted research on creating a Java-based, location-independent desktop environment—a forward-thinking project that anticipated the growing need for remote computing solutions. Based in Unionville, Connecticut, Jeff continues to be a driving force in the field of cloud computing, applying his extensive experience to help organizations harness the full potential of AWS technologies.
Jeff Harman is a Senior Prototyping Architect on the Amazon Web Services (AWS) Prototyping and Cloud Engineering team, he specializes in developing innovative solutions that leverage AWS’s cloud infrastructure to meet complex business needs. Jeff Harman is a seasoned technology professional with over three decades of experience in software engineering, enterprise architecture, and cloud computing. Prior to his tenure at AWS, Jeff held various leadership roles at Webster Bank, including Vice President of Platform Architecture for Core Banking, Vice President of Enterprise Architecture, and Vice President of Application Architecture. During his time at Webster Bank, he was instrumental in driving digital transformation initiatives and enhancing the bank’s technological capabilities. He holds a Master of Science degree from the Rochester Institute of Technology, where he conducted research on creating a Java-based, location-independent desktop environment—a forward-thinking project that anticipated the growing need for remote computing solutions. Based in Unionville, Connecticut, Jeff continues to be a driving force in the field of cloud computing, applying his extensive experience to help organizations harness the full potential of AWS technologies.
 Kosal Sen is a Design Technologist on the Amazon Web Services (AWS) Prototyping and Cloud Engineering team. Kosal specializes in creating solutions that bridge the gap between technology and actual human needs. As an AWS Design Technologist, that means building prototypes on AWS cloud technologies, and ensuring they bring empathy and value into the real world. Kosal has extensive experience spanning design, consulting, software development, and user experience. Prior to AWS, Kosal held various roles where he combined technical skillsets with human-centered design principles across enterprise-scale projects.
Kosal Sen is a Design Technologist on the Amazon Web Services (AWS) Prototyping and Cloud Engineering team. Kosal specializes in creating solutions that bridge the gap between technology and actual human needs. As an AWS Design Technologist, that means building prototypes on AWS cloud technologies, and ensuring they bring empathy and value into the real world. Kosal has extensive experience spanning design, consulting, software development, and user experience. Prior to AWS, Kosal held various roles where he combined technical skillsets with human-centered design principles across enterprise-scale projects.